
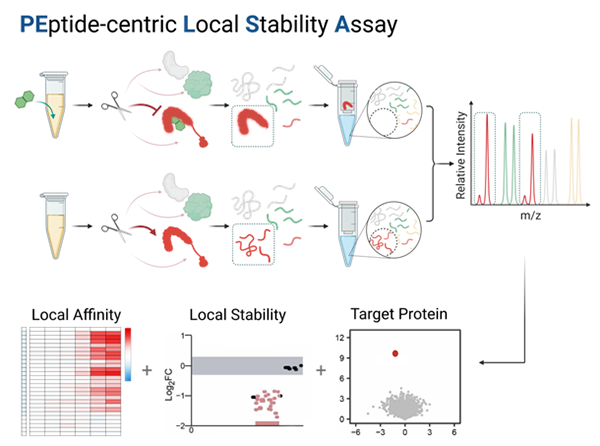
Identification of ligand target proteins and binding sites, determination
of local affinity
Supported by the National Natural Science Foundation of China (Grant No.
22204033, 92153302, 22137002), Ye Mingliang team of Dalian Institute of Chemical
Physics, Chinese Academy of Sciences cooperated with Luo Cheng team of Institute
of Materia Medica, Chinese Academy of Sciences, and made important progress in
the field of new methods for proteomic identification of ligand target proteins.
Related achievements are based on "A peptide centric local stability assay
enables proteome scale identification of the protein targets and binding regions
of diverse ligands and), published in Nature Methods recently. Paper link
https://www.nature.com/articles/s41592-024-02553-7 .
There are various ligand molecules in the intracellular and extracellular
environment, including metabolites, metal ions, nucleic acids, proteins, drugs,
etc. Their interactions with proteins affect the physiological and pathological
processes of organisms. Identifying the binding proteins and binding sites of
ligands can help reveal the mechanism of action of ligands in life activities,
which is of great significance for understanding complex life systems,
deciphering disease mechanisms, and promoting drug development. The traditional
ligand target proteomics identification method requires designing and optimizing
probe synthesis schemes that maintain ligand activity for different ligands,
which not only lacks broad-spectrum applicability but also makes it difficult to
identify ligand target proteins with weak interactions.
In response to these issues, the aforementioned team has developed a
peptide centered protein local stability assay (PELSA). This method does not
require chemical modification of ligands and does not depend on affinity size.
It can directly discover proteins that bind to ligands and undergo local
stability changes in complex systems such as cell lysate, thereby achieving
systematic analysis of ligand binding proteins, binding sites, and local
affinity. The identification sensitivity of this method for the target protein
of the model drug cyclosporine has increased by 12 times and 2.4 times compared
to existing similar technologies such as LiP-MS and TPP, respectively. In
addition, the dose-dependent PELSA method can determine local affinity, thereby
revealing the dynamic changes in the three-dimensional structure of proteins
after binding to ligands under physiological conditions. The team applied the
PELSA method to identify binding proteins of various ligands such as drugs,
metal ions, post-translational modified peptides, and antibodies, all of which
demonstrated highly sensitive target protein identification performance and
accurate binding region localization ability, proving that the PELSA method can
be used as a universal analysis platform without chemical modification of
ligands, and is widely applicable to target protein analysis of different
structural ligands.