
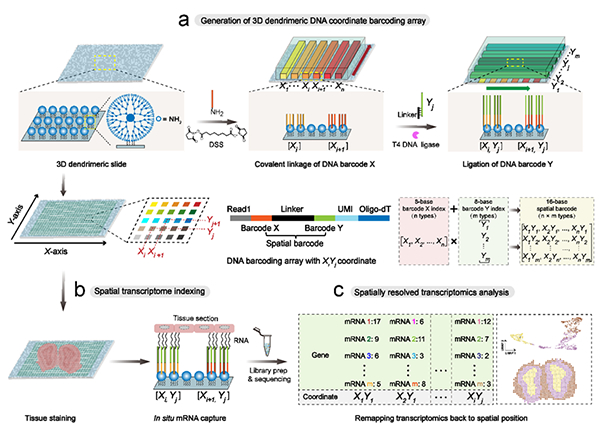
Schematic diagram of Decoder Seq technology for spatial transcriptome
sequencing
With the support of the National Natural Science Foundation of China (Grant
No. 21927806) and other grants, Professor Yang Chaoyong's team at Xiamen
University has made progress in the technology and application of spatial
transcriptome sequencing. The related research results, titled "Decoder seq
enhances mRNA capture efficiency in spatial RNA sequencing", were published in
the journal Nature Biotechnology. Paper link:
https://www.nature.com/articles/s41587-023-02086-y .
Space transcriptomics technology is a powerful tool for describing the
spatial expression patterns of genes within tissues, revealing cell composition,
spatial arrangement, and interactions. It has important application value in
major research areas such as organ structure, embryonic development,
neuroscience, life evolution, and human diseases. In recent years, sequencing
methods based on spatial barcode arrays have received great attention from
researchers due to their ability to provide preference free, high-throughput
spatial transcriptome analysis. However, this technology still faces bottlenecks
such as high cost, insufficient sensitivity, and low resolution.
The team utilized microfluidic assisted orthogonal coding strategy to
generate an array of high-density spatial barcodes on a three-dimensional (3D)
nano substrate, achieving low-cost, high-sensitivity, and high-resolution
spatial transcriptomics research. Firstly, the work constructed a 3D tree like
nano substrate, which increased the modification density of barcodes by about an
order of magnitude, thereby improving mRNA capture efficiency. Secondly, by
designing two microchannel chips with channels perpendicular to each other and
adjusting the number and width of channels on the chips, DNA coordinate barcode
lattices with different capture areas and spatial resolutions (50, 25, 15, and
10 μ m) were flexibly generated. Finally, the deterministic combination barcode
generated based on the orthogonal encoding strategy significantly reduces the
number of encoded DNA types, eliminating the need for decoding steps and
significantly reducing experimental costs. The sensitivity of Decoder seq with
near single-cell resolution (15 μ m) is as high as 40.1 mRNA molecules per μ m2,
which is much higher than other similar methods and achieves accurate mapping of
tissue single-cell spatial maps. Thanks to the significant improvement in
detection sensitivity, the team has discovered and confirmed for the first time
a new pattern of layered distribution of two Olfr genes, revealing spatial
immune heterogeneity in the microenvironment of different subtypes of renal cell
carcinoma tissues and identifying a set of genes associated with clinical
staging and prognosis evaluation of renal cell carcinoma.