
SnoRNAs are a highly conserved class of small non coding RNAs that primarily function by directing specific RNA modifications, such as 2 ′ - O-methylation and pseudouridylation. These modifications are crucial for ensuring the normal folding and function of ribosomal RNA (rRNA), small nuclear RNA (snRNA), etc. However, the function of snoRNAs goes far beyond that. They can also interact with various RNA molecules such as mRNA, participating in the regulation of gene expression, RNA stability, and translation efficiency in multiple biological processes.

Although the importance of snoRNA has been widely recognized, a comprehensive understanding of its target RNA has been limited.
On November 22nd, Professor He Chuan and Professor Pan Tao's team from the University of Chicago published a research paper titled "snoRNA facilitated protein secret revealed by transcriptome wide snoRNA target identification" in the top journal Cell.
This study developed an innovative chemical cross-linking based method that can comprehensively detect cellular RNA targets of snoRNA in human cells and mouse brain tissue, revealing thousands of previously unreported interactions between snoRNA and mRNA. These newly discovered interactions are not limited to the known snoRNA guided RNA modification sites, but also involve non classical functions beyond RNA modification, broadening our understanding of snoRNA functional diversity.

Research Highlights
SnoKARR seq reveals a wide range of snoRNA mRNA interactions across the transcriptome
Most snoRNA target mRNA interactions do not involve RNA modification
SNORA73 binds mRNA and 7SL RNA, acting as the 'adhesive' for the ternary complex
Why study snoRNA
SnoRNA mainly undergoes 2 ′ - O-methylation and pseudouridylation modifications in rRNA and snRNA through C/D-box snoRNA and H/ACA box snoRNA, while ScaRNA mainly guides these modifications in snRNA, stabilizes its structure, and regulates its interaction with pre messenger RNA (mRNA) to affect splicing. Each snoRNA provides one or two guiding sequences for rRNA modification, which helps regulate ribosome biogenesis, codon recognition, and ribosome ligand interactions.
Although about 80% of annotated snoRNAs in the human genome lack clear functions, some snoRNAs can regulate gene expression by affecting mRNA stability, editing, and splicing. More than 50 snoRNAs are dysregulated in over 12 types of cancer, genome deletions in the SNORD115/116 cluster are associated with Prader Willi symptoms (PWS), and mutations in SNORD118 lead to white matter encephalopathy. The mechanisms of these diseases are still not fully understood, mainly due to the lack of effective tools to identify the targets of snoRNAs and the modification status of target RNAs in the transcriptome.
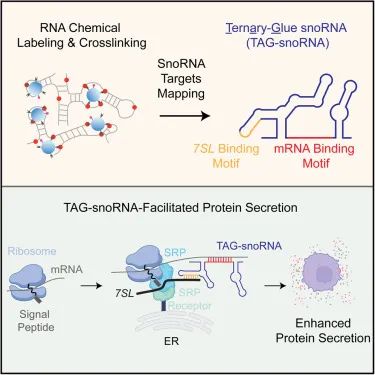
How to detect snoRNA targets?
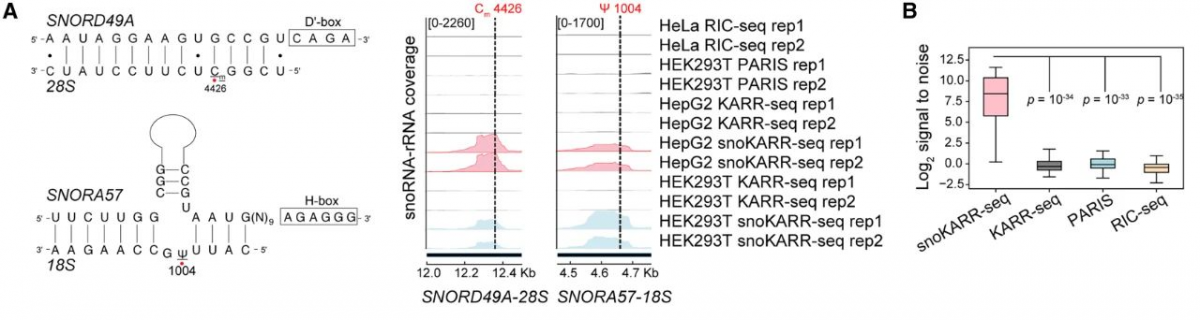
This study developed a method called KARR seq, which utilizes chemical crosslinkers to effectively capture physically adjacent RNA without relying on local RNA protein interactions. This method is based on the principle of ketoaldehyde assisted RNA-RNA interaction sequencing (KARR seq), using N3 ketoaldehyde to label guanosine in single stranded RNA (ssRNA), and using dibenzocyclooctane (DBCO) modified polyamine (PAMAM) dendrimers for RNA chemical cross-linking to capture RNA-RNA interactions. N3 ketoaldehyde can label various types of RNA, with an average distance of about 20 nucleotides (nt) between two labeling sites. In addition, this method introduces a cDNA enrichment step derived from snoRNA in the workflow, significantly improving the detection specificity of chimeric reads containing snoRNA in the final sequencing library.
SnoRNA73 promotes protein secretion
The research team used this method to identify thousands of snoRNA targets other than rRNA in different cell lines (HepG 2, MDA-MB-231, A549, PC 3, and HEK 293 T), and found that their interaction sites mostly occurred at non-traditional RNA modification sites, indicating that snoRNA may have other non classical regulatory effects.
So, the research team chose SNORA 73 with atypical structure, which is one of the most abundant H/ACA box snoRNAs in HepG2 cells and highly similar to SNORA 73 encoded by the human genome.
The research team detected a decrease in the expression levels of four target proteins by knocking down SNORA 73, and validated the non classical function of SNORA 73 in regulating protein secretion by binding to target mRNA through its mRNA binding motif (MBM) using spatial blockade technology.

Further research has found that SNORA73 forms a stable secondary structure with target mRNA through its non classical RNA binding sequence, and binds to the key RNA component 7SL RNA of signal recognition granules (SRP) to assemble into the "mRNAsnoRNA-7SL RNA" ternary complex.
SRP is a classic protein transport molecule responsible for recognizing signal peptides in newly formed peptide chains within cells and guiding them to the endoplasmic reticulum (ER) for secretion or membrane integration. SNORA73 acts as a "molecular glue" in this process, connecting SRP complexes with target mRNA through dual RNA-RNA interactions, significantly enhancing the binding efficiency between target mRNA and SRP, promoting endoplasmic reticulum transport of translation complexes, and thereby improving the transport and release of secreted proteins.
Experimental verification shows that knocking down the expression of SNORA73 leads to a significant decrease in the secretion level of its targeted secretory protein, while non targeted proteins are not affected. Subsequently, experimental results revealed that SNORA73 enhances protein secretion by promoting the transport of newly formed peptide chains, rather than by altering mRNA abundance or translation levels, further supporting the "glue" effect of SNORA73.

This study creatively proposed a more accurate snoRNA analysis method, providing a template for subsequent snoRNA related research. In the reference study, SNORA 73 was targeted and modified with green fluorescent protein (GFP) mRNA, significantly improving the secretion efficiency of GFP.
In the future, snoRNA may be artificially designed and modified to enhance the secretion efficiency of specific proteins, providing new ideas for the production of biological therapeutic proteins. The study of its mechanism also contributes to a deeper understanding of the molecular mechanisms of RNA modification and regulation, providing new targets and strategies for the diagnosis and treatment of various diseases.